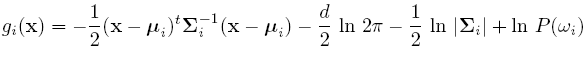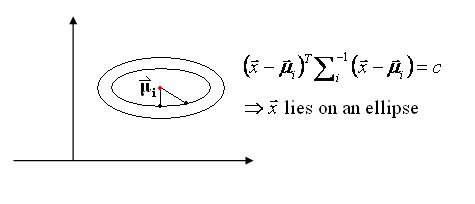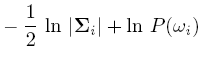| Line 98: | Line 98: | ||
If the Distribution Matrix is the same, then the second term goes off. | If the Distribution Matrix is the same, then the second term goes off. | ||
Special case 1: "Spherical clusters" in n-dimensional space. Equidistant points lie on a circle around the average values. | Special case 1: "Spherical clusters" in n-dimensional space. Equidistant points lie on a circle around the average values. | ||
| − | [[Image: ECE662_notes_5_OldKiwi.png | + | [[Image: ECE662_notes_5_OldKiwi.png]] |
| + | Figure 3 | ||
Final Note on Lecture 5: | Final Note on Lecture 5: | ||
| Line 104: | Line 105: | ||
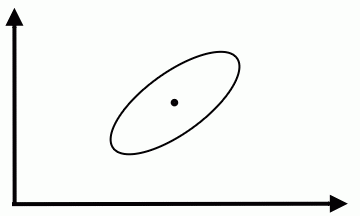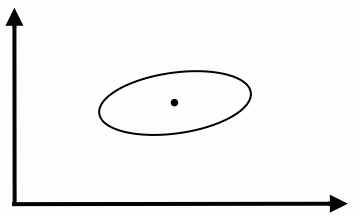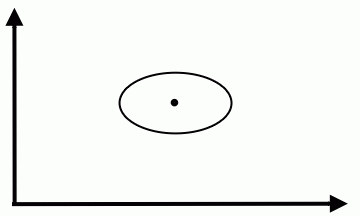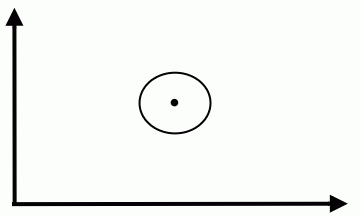
We could modify the coordinates of a class feature vector (as shown on the figure below) to take advantage of Special Case 1. But you should be aware that, in general, this can not be done for all classes simultaneously. | We could modify the coordinates of a class feature vector (as shown on the figure below) to take advantage of Special Case 1. But you should be aware that, in general, this can not be done for all classes simultaneously. | ||
| − | [[Image: change_coor1_OldKiwi.jpg | + | [[Image: change_coor1_OldKiwi.jpg]] |
| + | Figure 4 | ||
| − | [[Image: final_note_OldKiwi.gif | + | [[Image: final_note_OldKiwi.gif]] |
| + | Figure 5 | ||
== Lectures == | == Lectures == | ||
[http://balthier.ecn.purdue.edu/index.php/Lecture_1_-_Introduction 1] [http://balthier.ecn.purdue.edu/index.php/Lecture_2_-_Decision_Hypersurfaces 2] [http://balthier.ecn.purdue.edu/index.php/Lecture_3_-_Bayes_classification 3] | [http://balthier.ecn.purdue.edu/index.php/Lecture_1_-_Introduction 1] [http://balthier.ecn.purdue.edu/index.php/Lecture_2_-_Decision_Hypersurfaces 2] [http://balthier.ecn.purdue.edu/index.php/Lecture_3_-_Bayes_classification 3] | ||
[http://balthier.ecn.purdue.edu/index.php/Lecture_4_-_Bayes_Classification 4] [http://balthier.ecn.purdue.edu/index.php/Lecture_5_-_Discriminant_Functions 5] [http://balthier.ecn.purdue.edu/index.php/Lecture_6_-_Discriminant_Functions 6] [http://balthier.ecn.purdue.edu/index.php/Lecture_7_-_MLE_and_BPE 7] [http://balthier.ecn.purdue.edu/index.php/Lecture_8_-_MLE%2C_BPE_and_Linear_Discriminant_Functions 8] [http://balthier.ecn.purdue.edu/index.php/Lecture_9_-_Linear_Discriminant_Functions 9] [http://balthier.ecn.purdue.edu/index.php/Lecture_10_-_Batch_Perceptron_and_Fisher_Linear_Discriminant 10] [http://balthier.ecn.purdue.edu/index.php/Lecture_11_-_Fischer%27s_Linear_Discriminant_again 11] [http://balthier.ecn.purdue.edu/index.php/Lecture_12_-_Support_Vector_Machine_and_Quadratic_Optimization_Problem 12] [http://balthier.ecn.purdue.edu/index.php/Lecture_13_-_Kernel_function_for_SVMs_and_ANNs_introduction 13] [http://balthier.ecn.purdue.edu/index.php/Lecture_14_-_ANNs%2C_Non-parametric_Density_Estimation_%28Parzen_Window%29 14] [http://balthier.ecn.purdue.edu/index.php/Lecture_15_-_Parzen_Window_Method 15] [http://balthier.ecn.purdue.edu/index.php/Lecture_16_-_Parzen_Window_Method_and_K-nearest_Neighbor_Density_Estimate 16] [http://balthier.ecn.purdue.edu/index.php/Lecture_17_-_Nearest_Neighbors_Clarification_Rule_and_Metrics 17] [http://balthier.ecn.purdue.edu/index.php/Lecture_18_-_Nearest_Neighbors_Clarification_Rule_and_Metrics%28Continued%29 18] | [http://balthier.ecn.purdue.edu/index.php/Lecture_4_-_Bayes_Classification 4] [http://balthier.ecn.purdue.edu/index.php/Lecture_5_-_Discriminant_Functions 5] [http://balthier.ecn.purdue.edu/index.php/Lecture_6_-_Discriminant_Functions 6] [http://balthier.ecn.purdue.edu/index.php/Lecture_7_-_MLE_and_BPE 7] [http://balthier.ecn.purdue.edu/index.php/Lecture_8_-_MLE%2C_BPE_and_Linear_Discriminant_Functions 8] [http://balthier.ecn.purdue.edu/index.php/Lecture_9_-_Linear_Discriminant_Functions 9] [http://balthier.ecn.purdue.edu/index.php/Lecture_10_-_Batch_Perceptron_and_Fisher_Linear_Discriminant 10] [http://balthier.ecn.purdue.edu/index.php/Lecture_11_-_Fischer%27s_Linear_Discriminant_again 11] [http://balthier.ecn.purdue.edu/index.php/Lecture_12_-_Support_Vector_Machine_and_Quadratic_Optimization_Problem 12] [http://balthier.ecn.purdue.edu/index.php/Lecture_13_-_Kernel_function_for_SVMs_and_ANNs_introduction 13] [http://balthier.ecn.purdue.edu/index.php/Lecture_14_-_ANNs%2C_Non-parametric_Density_Estimation_%28Parzen_Window%29 14] [http://balthier.ecn.purdue.edu/index.php/Lecture_15_-_Parzen_Window_Method 15] [http://balthier.ecn.purdue.edu/index.php/Lecture_16_-_Parzen_Window_Method_and_K-nearest_Neighbor_Density_Estimate 16] [http://balthier.ecn.purdue.edu/index.php/Lecture_17_-_Nearest_Neighbors_Clarification_Rule_and_Metrics 17] [http://balthier.ecn.purdue.edu/index.php/Lecture_18_-_Nearest_Neighbors_Clarification_Rule_and_Metrics%28Continued%29 18] | ||
Revision as of 21:44, 23 March 2008
LECTURE THEME : - Discriminant Functions
Discriminant Functions: one way of representing classifiers
Given the classes $ \omega_1, \cdots, \omega_k $
The discriminant functions $ g_1(x),\ldots, g_K(x) $ such that $ g_i(x) $ n-dim S space $ \rightarrow \Re $
which are used to make decisions as follows:
decide $ \omega_i $ if $ g_i(x) \ge g_j(x), \forall j $
Note that many different choices of $ g_i(x) $ will yield the same decision rule, because we are interested in the order of values of $ g_i(x) $ for each x, and not their exact values.
For example: $ g_i(x) \rightarrow 2(g_i(x)) $ or $ g_i(x) \rightarrow ln(g_i(x)) $
In other words, we can take $ g_i(x) \rightarrow f(g_i(x)) $ for any monotonically increasing function f.
Relation to Bayes Rule
e.g. We can take $ g_i(\mathbf(x)) = P(\omega_i|\mathbf(x)) $
then $ g_i(\mathbf(x)) > g_j(\mathbf(x)), \forall j \neq i $
$ \Longleftrightarrow P(w_i|\mathbf(X)) > P(w_j|\mathbf(X)), \forall j \neq i $
OR we can take
$ g_i(\mathbf(x)) = p(\mathbf(x)|\omega_i)P(\omega_i) $
then $ g_i(\mathbf(x)) > g_j(\mathbf(x)), \forall j \neq i $
$ \Longleftrightarrow g_i(\mathbf(x)) = ln(p(\mathbf(x)|\omega_i)P(\omega_i)) = ln(p(\mathbf(x)|\omega_i))+ln(P(\omega_i) $
OR we can take
$ g_i(\mathbf(x)) = ln(p(\mathbf(x)|\omega_i)P(\omega_i)) = ln(p(\mathbf(x)|\omega_i))+ln(P(\omega_i) $
We can take any $ g_i $ as long as they have the same ordering in value as specified by Bayes rule.
Some useful links:
- Bayes Rule in notes: https://engineering.purdue.edu/people/mireille.boutin.1/ECE301kiwi/Lecture4
- Bayesian Inference: http://en.wikipedia.org/wiki/Bayesian_inference
Relational Decision Boundary
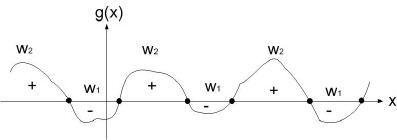
Ex : take two classes $ \omega_1 $ and $ \omega_2 $
$ g(\vec x)=g_1(\vec x)-g_2(\vec x) $
decide $ \omega_1 $ when $ g(\vec x)>0 $
and $ \omega_2 $ when $ g(\vec x)<0 $
when $ g(\vec x) = 0 $, you are at the decision boundary ( = hyperplane)
$ \lbrace \vec x | \vec x \;\;s.t \;\;g(\vec x)=0\rbrace $ is a hypersurface in your feature space i.e a structure of co-dimension one less dimension than space in which $ \vec x $ lies
Discriminant function for the Normal Density
Suppose we assume that the distribution of the feature vectors is such that the density function p(X|w) is normal for all i.
Eg: Length of hair among men is a normal random variable. Same for hairlength in women. Now we have:
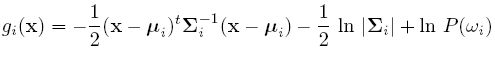
For simplicity, add n/2ln(2pi) to all gi's. Then we get:
If classes are equally likely, then we can get rid of the first term.
If the Distribution Matrix is the same, then the second term goes off.
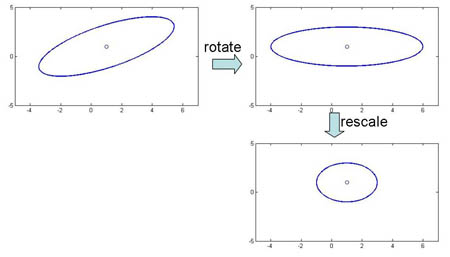
Special case 1: "Spherical clusters" in n-dimensional space. Equidistant points lie on a circle around the average values.
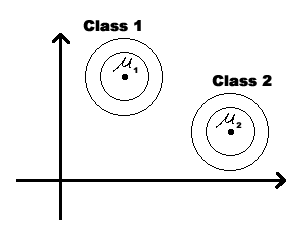 Figure 3
Figure 3
Final Note on Lecture 5:
We could modify the coordinates of a class feature vector (as shown on the figure below) to take advantage of Special Case 1. But you should be aware that, in general, this can not be done for all classes simultaneously.